Short-read sequencing has revolutionized the field of biomedical research. Compared to traditional methods, it’s faster and more cost-effective.
Short reads are effective for applications aimed at counting the abundance of specific sequences, identifying variants within otherwise well-conserved sequences, or profiling the expression of particular transcripts.
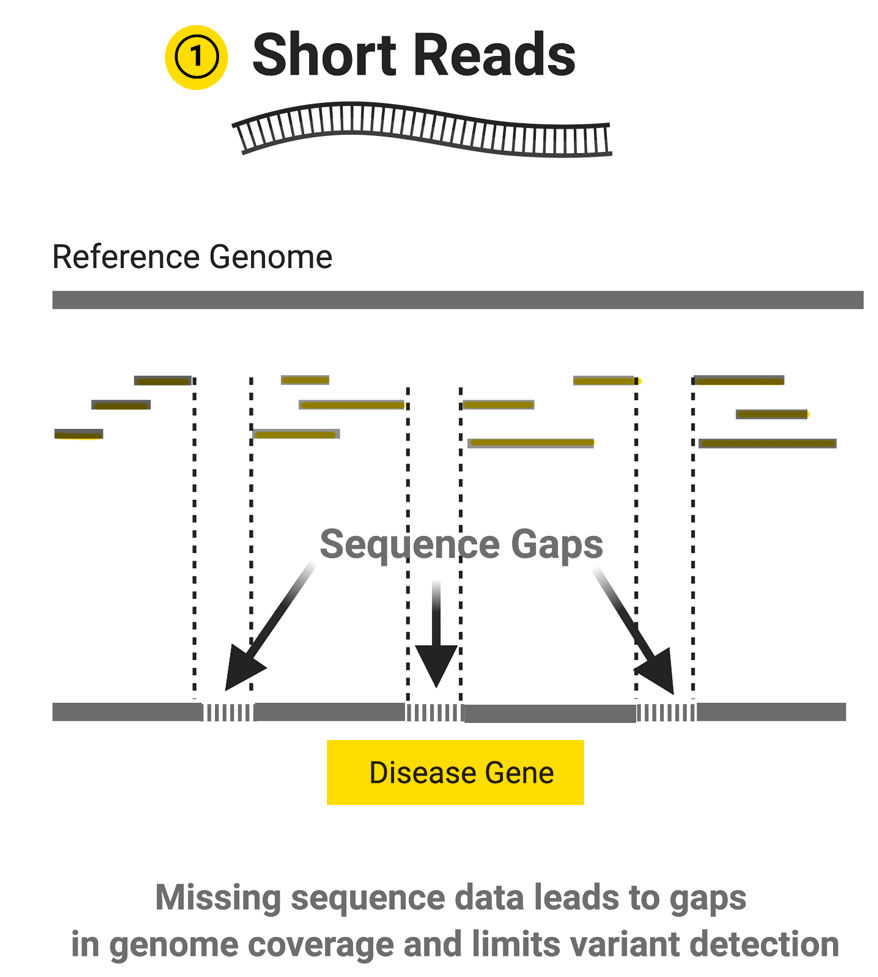
Image source: https://www.hudsonalpha.org/piecing-together-the-genome-the-long-and-short-of-it-all/
Here are some of the short-read sequencing technologies making waves in the scientific community:
📍Illumina: They use single-stranded DNA-binding proteins to amplify DNA and then add fluorescent-labeled deoxynucleoside triphosphates to bridge the amplified DNA template.
📍454 pyrosequencing: These folks take advantage of clonal amplification through emulsion PCR. They create microbead-bound DNA clones in separate water-in-oil droplets to avoid mix-ups. DNA polymerase is added, and pyrophosphate release is monitored to track nucleotide incorporation.
📍Ion Torrent: Emulsion PCR also comes into play here for clonal amplification. The DNA clones are trapped on microbeads, and a pH sensor detects the release of protons as deoxynucleoside triphosphates are incorporated.
📍SOLiD: Emulsion PCR strikes again! DNA templates are bound to microbeads this time and hybridized with adapters using DNA ligase.
While these technologies use different chemistries, they all generate reads that are at most, a few hundred bases long. Even still, the reads are high quality, and there are a lot of them – anywhere from a few million to hundreds of billions depending on the sequencer. This means researchers can get higher coverage of their genomes or targets of interest and enables high-confidence SNP and mutation calling.
Long-read sequencers are able to generate reads with much longer lengths – anywhere from a few thousand to hundreds of thousands of bases. These longer reads allow researchers to more easily identify complex structural variations such as large insertions/deletions, inversions, repeats, duplications, and translocations.
This sequencing technology can also be used to phase SNPs into haplotypes, build scaffolds for de novo assembly and resolve splicing events in full-length cDNA.
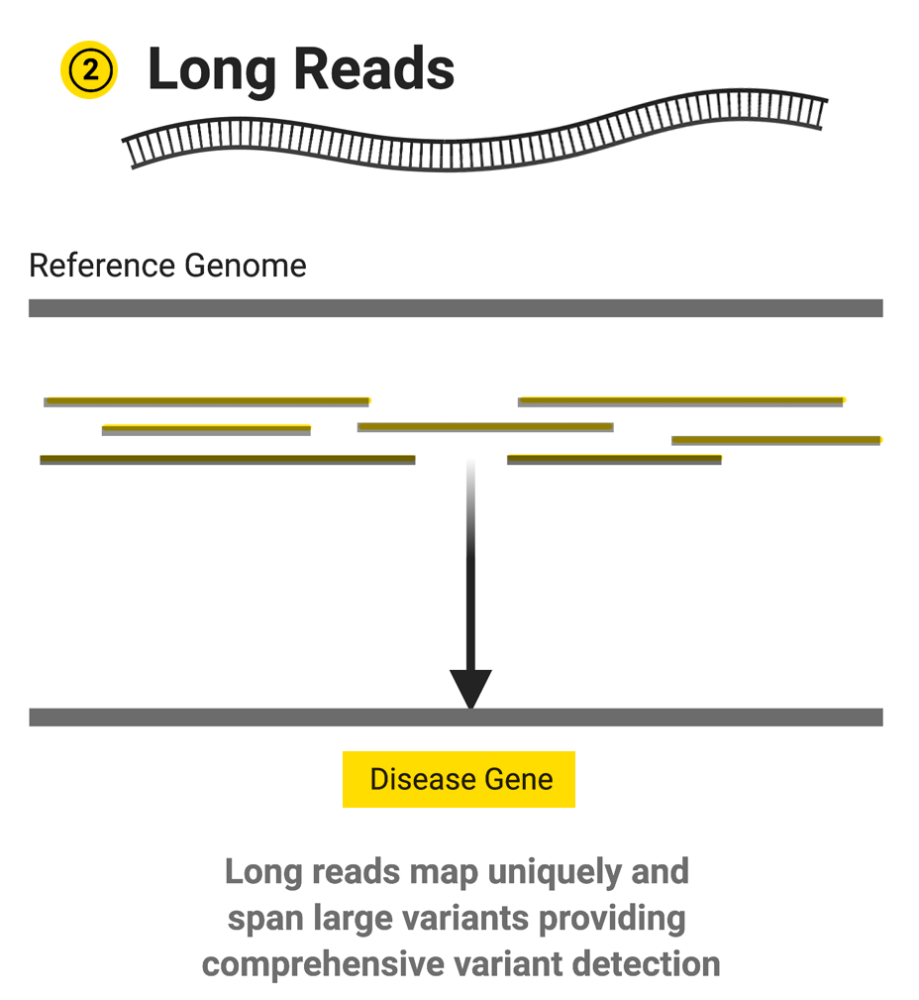
Image source: https://www.hudsonalpha.org/piecing-together-the-genome-the-long-and-short-of-it-all/
Long-read instruments have been on the market for the past decade but the lower yield, higher error rate, and higher costs of the instruments have kept them from being more widely adopted.
An additional downside is that the accuracy per read can be much lower than that of short-read sequencing.
The high error rate of nanopore technology is largely due to the inability to control the speed of the DNA molecules through the pore – these are systematic errors. Errors in SMRT sequencing are completely random. These can be reduced by circular consensus sequencing; a method that allows DNA to pass through the zero-mode waveguide chip several times, generating highly accurate reads of at least 99.8%, similar to NGS platforms.
The stars of long-read sequencing:
📍Pacific Biosciences (PacBio): They developed a nifty sequencer called Single Molecule Real-Time (SMRT). This bad boy can generate reads exceeding 10,000 bases in less than two hours. The sequencing action takes place on a zero-mode waveguide chip, where DNA polymerase is fixed at the bottom. With the help of adapters, the DNA molecule forms a circular single-stranded structure. Complementary strands are sequenced, and fluorescence measurements reveal the nucleotides. PacBio’s latest creation, the Revio system, brings even more HiFi data and human genomes at scale for less than $1,000!
📍Oxford Nanopore Technologies: Prepare to be amazed, because this platform can produce reads of up to a staggering 1 million base pairs! They achieve this by threading DNA molecules through a nanopore, a specially engineered channel in a biological membrane. The electrical current across the channel changes depending on the specific nucleotide passing through.
The Best of Both Worlds
Some challenges can be resolved by using a combination of short-read and long-read technologies.
10x Genomics has developed a method called ‘Linked-Reads’, which essentially provides long-range information from short-read sequencing data. It uses molecular barcodes to tag short-reads that originate from the same long DNA fragment, making it possible to link all of the short-reads together.
Linked reads allow for a significantly increased physical coverage with only a slight increase of standard sequencing, ultimately providing better access to typically inaccessible parts of the genome and overcoming several of the limitations of short-read sequencing.

