Next-generation sequencing (NGS) is a powerful tool that sequences genomes or targeted regions of DNA or RNA. NGS has revolutionized molecular biology and has allowed significant progress in understanding the genetic basis of disease, as well as to develop new treatments. NGS has also enabled the development of personalized medicine, which involves the use of a patient’s genetic information to tailor treatments to their specific needs.
When the first human genome was sequenced, in 1987, it cost $2.7 billion, required international collaboration, and still took a decade to complete. Now, 30 years later it can be done for $1000, without collaboration, and in just 3 hours!
This has impacted numerous research fields, including:
- Clinical Diagnostics: Next-generation sequencing NGS is highly diverse and ranges from the detection of novel microbial agents that cause epidemics to detect heterogeneous mutations that cause complex disorders like autism, connective tissue disorders, and cardiomyopathies.
“The rate of progress is stunning. As costs continue to come down, we are entering a period where we are going to be able to get the complete catalog of disease genes. This will allow us to look at thousands of people and see the differences among them, to discover critical genes that cause cancer, autism, heart disease, or schizophrenia.” – Eric Lander, founding director of the Broad Institute of MIT and Harvard and principal leader of the Human Genome Project.
- Vaccines: From investigating the origin of the SARS-CoV-2 virus to the rapid development of vaccines, sequencing has been a tremendous help during the COVID-19 pandemic.
- Agriculture: NGS is used to develop new crop varieties that are more resistant to diseases and pests, and to identify genetic markers associated with desirable traits such as yield or flavor.
- Evolution: NGS has been used to compare the human genome with the other species, and decipher how closely or distantly we may be related to them. Mutations are vital in evolution. NGS allows scientists to study how mutations have accumulated throughout evolution.
- Forensics: Next-generation sequencing (NGS) now gives forensic testing the potential to deliver extended capabilities, higher resolution, and greater efficiency—leading to more reliable results. This enables identifying missing persons, victims of mass disasters, unidentified remains, complex familial relationships, and promising investigative leads.
Although there are many different machines on the market each with different types of sequencing methods, they all use the same 4 basic steps:
- Nucleic Acid Extraction: NGS can be performed on any sample that yields DNA or RNA, including cell cultures, fresh-frozen tissues, formalin-fixed paraffin-embedded (FFPE) tissues, blood, saliva, and bone marrow.
- Library preparation: A library is a collection of similarly sized DNA fragments with adapter sequences – a short piece of DNA that allows the fragments of DNA to be identified. This boosts the efficiency of the library preparation process.
- Sequencing: Parallel sequencing is performed using an NGS platform. The library is loaded onto the sequencer which then “reads” the nucleotides one by one.
- Data Analysis: Each machine provides the raw data at the end of the sequencing run. The newly identified sequence reads are aligned to a reference genome.
Even though current NGS platforms are very accurate, they are still prone to error. Even at accuracies greater than 99%, a sequence generated may contain incorrect nucleotides. This means that if a machine’s accuracy is 99%, one base pair is misread out of 100. Since NGS platforms generate high amounts of output (human genome 3.2 billion base pairs long), these errors can add up quickly.
Check-list for High-Quality Nucleic Acids
- Absorbance Value: The purity of a nucleic acid sample is typically expressed as an A260/280 value, which can be determined using a spectrophotometer. “Pure” DNA generally reports an A260/280 reading of 1.8, whereas RNA is closer to 2.0. Values different from the aforementioned state presence of contaminants (including protein, EDTA, Phenol, etc.).
- High Yield: The result obtained must suffice NGS workflow requirements.
- Intact State: Concentrations and pH of buffers may cause shearing of nucleic acids.
Quickpure Genomic DNA Isolation Kit
The Quickpure Blood gDNA Extraction Kit from Cambrian Bioworks is designed for rapid, high-throughput isolation of high-quality genomic DNA. The kit uses CamBeads magnetic bead technology, ensuring reproducible recovery of PCR-ready DNA suitable for a broad range of applications.

Features:
- Automated – 96 DNA isolations in less than 20 minutes
- Versatile – Single kit for multiple samples types specialized buffer system
- Optimized for higher yields
- Convenient – Custom Protocols are available
- High-quality support– Our application scientists will help you onboard the new protocol into your workflow
Performance Proof
Three blood samples from different sources were collected in an EDTA Vacutainer and processed using Cambrian quickpure DNA isolation kit on a kingfisher flex system.
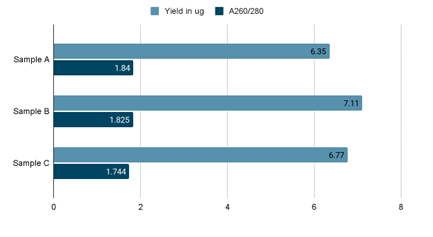

If you’re a lab, a biotechnology or a genomics company, or anyone who thinks that our Blood gDNA kit could be useful to you, reach out to us for a free sample.
To get more information, contact us: +91 90356 74375

